UQ PhD student develops SMART tool against antibiotic resistant superbugs
Antibiotic resistant superbugs are a serious health threat worldwide and are associated with high morbidity and mortality. Rapid molecular detection of superbugs is important for infection control and guiding appropriate treatment decisions.
To understand how rapid molecular assays are applied; a typical scenario might be, physicians commence empirical therapy for a sick patient if they suspect bacterial infection, based on clinical signs and symptoms. At the same time, diagnostic samples (blood and urine cultures) are collected to identify the causative organism and potential antibiotic resistance.
From a laboratory point of view, traditional culture-based diagnostic assays that require sufficient bacterial growth can be slow. By contrast, culture-independent methods, such as molecular diagnostic assays, could quicken turnaround times and provide early actionable information to initiate targeted antibiotic therapy. This can be an essential element for improving outcomes in patients with life-threatening infections.
Despite the many advantages of well-established assays, they can be a quite problematic when applied in low income settings and/or where the care of patient is critical. Reasons for this include; turnaround times can be lengthy, results depend on instrument and expertise analysis, and they can be expensive to perform, due to high purchasing costs of instruments and/or commercial kits.
Here is where my PhD project plays a role. I address the above limitations and improve diagnostics by providing healthcare workers with innovative tools that could potentially satisfy the World Health Organisation’s (WHO) ASSURED criteria (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, deliverable). These tools can also be used for suitable point-of-care (POC) testing in low income settings.
I call them SMART methods; Simple, Molecular Antibiotic Resistance Tests that offer assays to perform and interpret results without the need for expertise and potentially instruments.
As part of my PhD, I am targeting and tackling major antibacterial resistance threats identified by the WHO and USA Centre for Disease Prevention and Control (CDC). So far, I have successfully developed methods to detect a family of antibiotic resistance genes called carbapenemases. These developed assays can provide results much faster than conventional molecular assays with results available in minutes, rather than hours or days. Initial results are very promising with key performance characteristics like sensitivity and specificity comparing favourably with established laboratory-based tests.
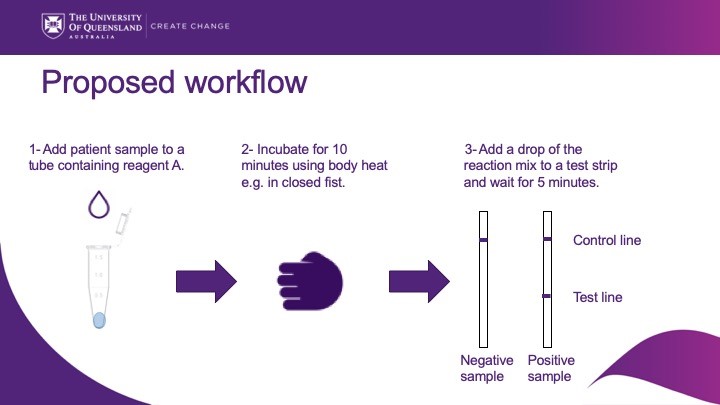
The key advantages of the developed assays, apart from being very fast, is that they utilise a simple lateral flow-based detection system similar to a home pregnancy test. This enables positive samples to be readily distinguished from negative samples on the presence or absence of bands.
The key difference with a pregnancy test is that for antibiotic resistance genes we must first ‘amplify’ the target so that we can generate sufficiently strong bands that can be seen by the naked eye. This amplification step requires heat, and this is where we are looking to incubate the reaction tubes using body heat.
There’s still a lot to be done, but I am enjoying conducting the various experiments to enhance the performance and speed of these methods. I’m looking forward seeing these assays trialled in a clinic setting.
Abdulrahman’s PhD is sponsored by King Abdul-Aziz University (KAU), and his PhD project is funded by The Children`s Hospital Foundation (CHF) with researchers; Dr. Adam Irwin, Dr. Hosam Zowawi, A/Prof Joanne Macdonald, Dr. Claire Heney, Dr. Patrick Harris, A/Prof David Whiley, Prof. David Paterson and Post-doc Nicole Ertl, as well as industry partner Biocifer Pty Ltd Australia.

